Stress eating: hypothalamic control over dopamine system drives bingeing
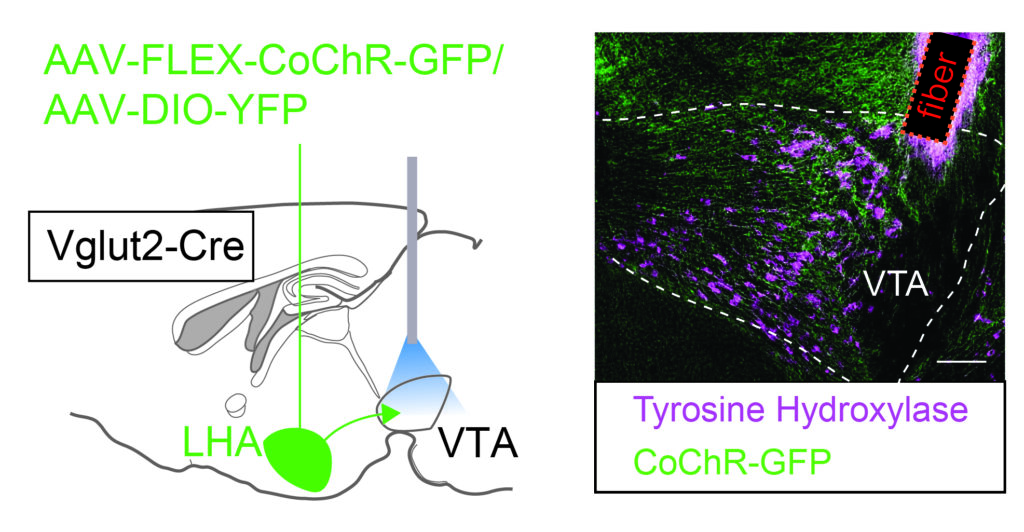
Does stress eating sound familiar? Stress can increase the intake of high caloric food, which can contribute to obesity and eating disorders, but the neurobiology underlying this process is not clear. In a new paper out in Nature Communications the lab of Frank Meye describes how stress changes synaptic strength from the Lateral Hypothalamus (LHA) to the Ventral Tegmental Area (VTA) midbrain dopamine system to drive binge eating behavior.
First author Louisa Linders observed in mice that social stress resulted in binge-like intake of high caloric fat. Calcium recordings done by Lefkothea Patrikiou and Evelien Schut showed that LHA glutamatergic neurons controlling the VTA were responsive to such social stress, as well as to fat intake. Using patch-clamp electrophysiological approaches, Louisa went on to show that social stress strengthened the glutamatergic synaptic connection from the LHA to the VTA. Optogenetic tweaking of the strength of these LHA-VTA synapses proved critical in regulating whether stress eating took place. Overall, this paper highlights an important causal role of stress-induced plasticity in the synaptic connection from the lateral hypothalamus to the midbrain dopamine reward system for stress-driven food intake.
“I’m excited that we can share our findings on how stress changes synaptic function to causally contribute to stress eating.” Louisa Linders
The work was performed in the Translational Neuroscience Dept of the UMC Utrecht Brain Center. Check out the tweet by Frank Meye as well.
We look forward to seeing the upcoming work from the Meye group!
Studying synapses in specific neural circuits
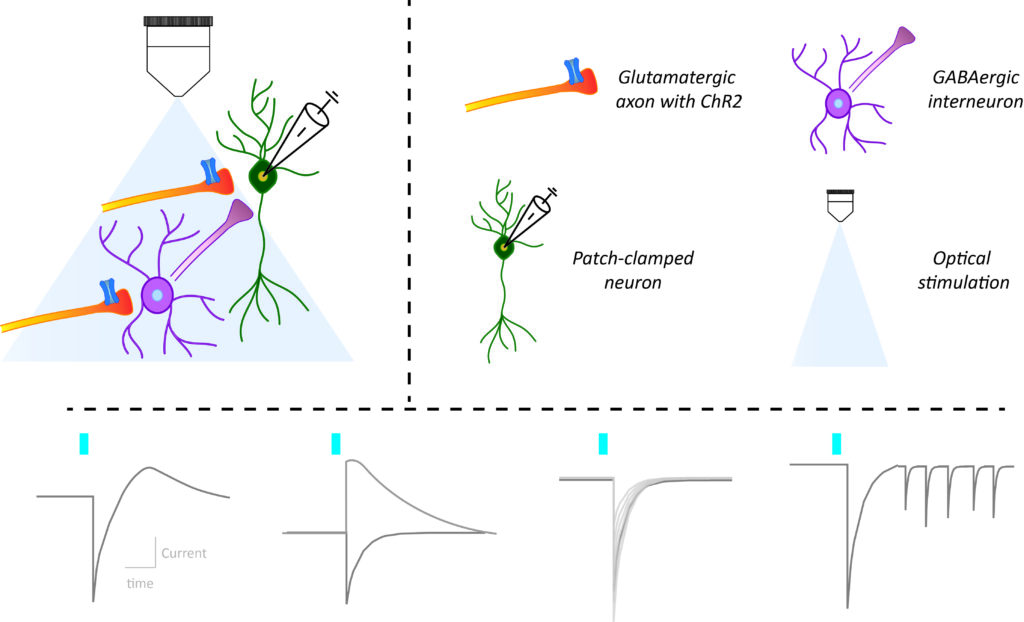
In recent years the combination of brain slice electrophysiological recordings of neurons in response to optogenetic stimulation of their input, has become an indispensable staple to probe the function of neural circuits. The large amounts of distinct metrics that one can obtain this way are both informative but also daunting to the newcomer. In a recent review paper, Frank Meye’s team summarized the methodological state-of-the-art for this approach. The paper featured several team members including co-first authors Laura Supiot and Louisa Linders. In particular, the authors reviewed the rationale behind the different metrics used to study synaptic connectivity and changes in synaptic strength. They compiled a guide for the implementation of these methodologies in practice and discussed future directions to decipher neural circuits.
“We aimed to provide the rationale for distinct electrophysiological synaptic metrics, as well as practically explain how to obtain them.” Laura Supiot
Also see the tweet by Frank Meye. Congratulations to the whole team! 🎉
Leptin targets spatially diverse neurons


Leptin is a hormone that is secreted by fat and signals the need to stop eating and increase energy expenditure via leptin receptors (LepR). Various hunger and reward centers in the brain contain different LepR expressing neurons. The primary leptin center is the well-studied arcuate nucleus. Other hypothalamic nuclei are less abundant in leptin receptor but also essential in encoding leptin’s actions. The composition of these LepR neurons have not been well understood.
In their recently published paper in Science Reports, Nefeli Kakava and colleagues from the UMC Utrecht Brain Center explore the scarce LepR population in the lateral hypothalamus. This population is known for its effects on food intake and food reward, and may be defective in eating disorders. The authors successfully capture the transcriptome of these neurons using TRAP-Seq. Exploration of their molecular profile confirms the expression of diverse neuropeptides and receptors. Microscopy analysis reveal their diverse spatial expression patterns. Moreover, they unravel new markers that could have significant role in energy balance. They also explore what is the transcriptional response of these neurons to energy deficit.
“I am excited that we have successfully managed to capture RNA from this very rare albeit significant population of leptin responsive cells and hope our findings inspire new research”
Nefeli obtained her PhD in 2020 as part of the Adan lab, where she developed viral vector tools to target and manipulate the activity of brain cells involved in food reward and energy balance. In collaboration with the Basak lab, she has profiled the hypothalamic LepR cells using TRAP-Seq and single cell genomics techniques.

Characterising social stress responsive ventral tegmental area neurons

In this study published at Frontiers in Behavioral Neuroscience, Ioannis Koutlas and colleagues of the Meye lab, use the expression of immediate-early genes to characterize social-stress activated neuronal subsets in the ventral tegmental area. They show that cells of different molecular identities (dopaminergic, GABAergic, glutamatergic and combinatorial neurons) that are dispersed throughout the entire VTA are activated by a social stress episode. Furthermore, they validate the use of targeted recombination in active populations (TRAP2) to capture this VTA stress-activated neuronal ensemble and make it tractable for further manipulations. Finally, the use of TRAP2 allowed them to look into intrinsic electrophysiological properties of these neurons and show that stress activated VTA cells are more excitable than neighbouring cells that were not activated by stress.

“I am very excited that this work is published. It gives insight on stress-encoding VTA neuronal populations and provides us with the tools to answer further exciting questions” – Ioannis
Cellular diversity of the developing mouse Habenula
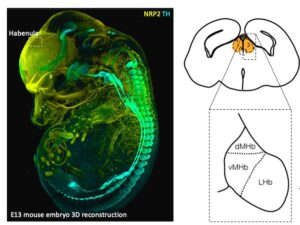
The habenula (Hb) plays a key role in processing reward information and mediating aversive responses to negative stimuli. In the recent issue of Cell Reports, Lieke van de Haar and colleagues have revealed how the cellular diversity of the mouse habenula is formed during development. In the work title “Cellular diversity of developing habenula may illuminate risk for human psychiatric disorders”, Lieke used single cell RNA sequencing, a technique that allows quantification of gene expression in thousands of cells simultaneously.
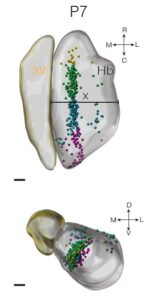 Reconstruction of lineage trajectories using computational tools revealed paths to habenula cell types. Intersectional genetics experiments by Oxana Garritsen showed that these include a physiologically distinct neuronal cell type that innervate the dorsal IPN which can be identified by Cartpt expression. Work by Lieke and Youri Adolfs using iDISCO and light sheet microscopy have shown that these cells localize to the medial habenula. The ePhys work by Danai Riga of the Meye lab found that these Cartpt+ neurons are electrophysiologically different from surrounding neurons. Finally, MAGMA analysis by Juliska Boer, who did her bachelor internship under the supervision of Lieke, revealed that developing cell populations align with human genetic risk loci of psychiatric disorders, such as the major depressive disorder.
Reconstruction of lineage trajectories using computational tools revealed paths to habenula cell types. Intersectional genetics experiments by Oxana Garritsen showed that these include a physiologically distinct neuronal cell type that innervate the dorsal IPN which can be identified by Cartpt expression. Work by Lieke and Youri Adolfs using iDISCO and light sheet microscopy have shown that these cells localize to the medial habenula. The ePhys work by Danai Riga of the Meye lab found that these Cartpt+ neurons are electrophysiologically different from surrounding neurons. Finally, MAGMA analysis by Juliska Boer, who did her bachelor internship under the supervision of Lieke, revealed that developing cell populations align with human genetic risk loci of psychiatric disorders, such as the major depressive disorder.
“The habenula plays an important role in negative experiences, and is located dorsally to the thalamus and ventrally to the hippocampus…. we were interested in how the cells of the habenula are born and mature.” Lieke
Lieke performed her PhD work at the Pasterkamp lab of our Translational neuroscience department of the UMC Utrecht Brain Center. This work is a fine example of internal collaborations in the department which connect molecular, computational, genetic and electrophysiological techniques.